Bayesian Hierarchical Modelling
In this last practical we will consider the analysis of Bayesian hierarchical models. As explained in the previous lecture, hierarchical models provide a convenient tool to define models so that the different sources of variation in the data are clearly identified. Bayesian inference for highly structured hierarchical models can be difficult and may require the use of Markov chain Monte Carlo methods.
However, packages such as BayesX and INLA,
to mention just two, provide a very convenient way to fit and make
inference about certain types of Bayesian hierarchical models. We will
continue using BayesX, although you can try out using INLA
if you look at the optional, additional material in Practical 8.
Linear Mixed Models
Linear mixed models were defined in the lecture as follows:
Here, represents observation in group , are a vector of covariates with coefficients , i.i.d. random effects and a Gaussian error term. The distribution of the random effects is Gaussian with zero mean and precision .
Multilevel Modelling
Multilevel models are a particular type of mixed-effects models in which observations are nested within groups, so that group effects are modelled using random effects. A typical example is that of students nested within classes.
For the next example, the nlschools data set (in package
MASS) will be used. This data set records data about
students’ performance (in particular, about a language score test) and
other variables. The variables in this data set are:
lang, language score test.IQ, verbal IQ.class, class ID.GS, class size as number of eighth-grade pupils recorded in the class.SES, social-economic status of pupil’s family.COMB, whether the pupils are taught in the multi-grade class with 7th-grade students.
The data set can be loaded and summarised as follows:
## lang IQ class GS SES
## Min. : 9.00 Min. : 4.00 15580 : 33 Min. :10.00 Min. :10.00
## 1st Qu.:35.00 1st Qu.:10.50 5480 : 31 1st Qu.:23.00 1st Qu.:20.00
## Median :42.00 Median :12.00 15980 : 31 Median :27.00 Median :27.00
## Mean :40.93 Mean :11.83 16180 : 31 Mean :26.51 Mean :27.81
## 3rd Qu.:48.00 3rd Qu.:13.00 18380 : 31 3rd Qu.:31.00 3rd Qu.:35.00
## Max. :58.00 Max. :18.00 5580 : 30 Max. :39.00 Max. :50.00
## (Other):2100
## COMB
## 0:1658
## 1: 629
##
##
##
##
## The model to fit will take lang as the response variable
and include IQ, GS, SES and
COMB as covariates (i.e., fixed effects). This model can
easily be fit with BayesX (using the R2BayesX
package) as follows:
## Call:
## bayesx(formula = lang ~ IQ + GS + SES + COMB, data = nlschools)
##
## Fixed effects estimation results:
##
## Parametric coefficients:
## Mean Sd 2.5% 50% 97.5%
## (Intercept) 9.7059 1.0630 7.6150 9.7073 11.7042
## IQ 2.3878 0.0723 2.2494 2.3892 2.5301
## GS -0.0263 0.0246 -0.0735 -0.0261 0.0227
## SES 0.1483 0.0143 0.1195 0.1488 0.1754
## COMB1 -1.6901 0.3123 -2.3009 -1.6999 -1.0776
##
## Scale estimate:
## Mean Sd 2.5% 50% 97.5%
## Sigma2 48.0652 1.4518 45.2863 48.0170 51.07
##
## N = 2287 burnin = 2000 method = MCMC family = gaussian
## iterations = 12000 step = 10Note that the previous model only includes fixed effects. The data
set includes class as the class ID to which each student
belongs. Class effects can have an impact on the performance of the
students, with students in the same class performing similarly in the
language test.
Very conveniently, Bayesx can include random effects in
the model by adding a term in the right hand side of the formula that
defined the model. Specifically, the term to add is
sx(class, bs = "re") (see code below for the full model).
This will create a random effect indexed over variable
class and which is of type re, i.e., it
provides random effects which are independent and identically
distributed using a normal distribution with zero mean and precision
- there are clear similarities with a residual terms here (although that
is at the observation level, rather than the group level).
Before fitting the model, the between-class variability can be explored by means of boxplots:
boxplot(lang ~ class, data = nlschools, las = 2)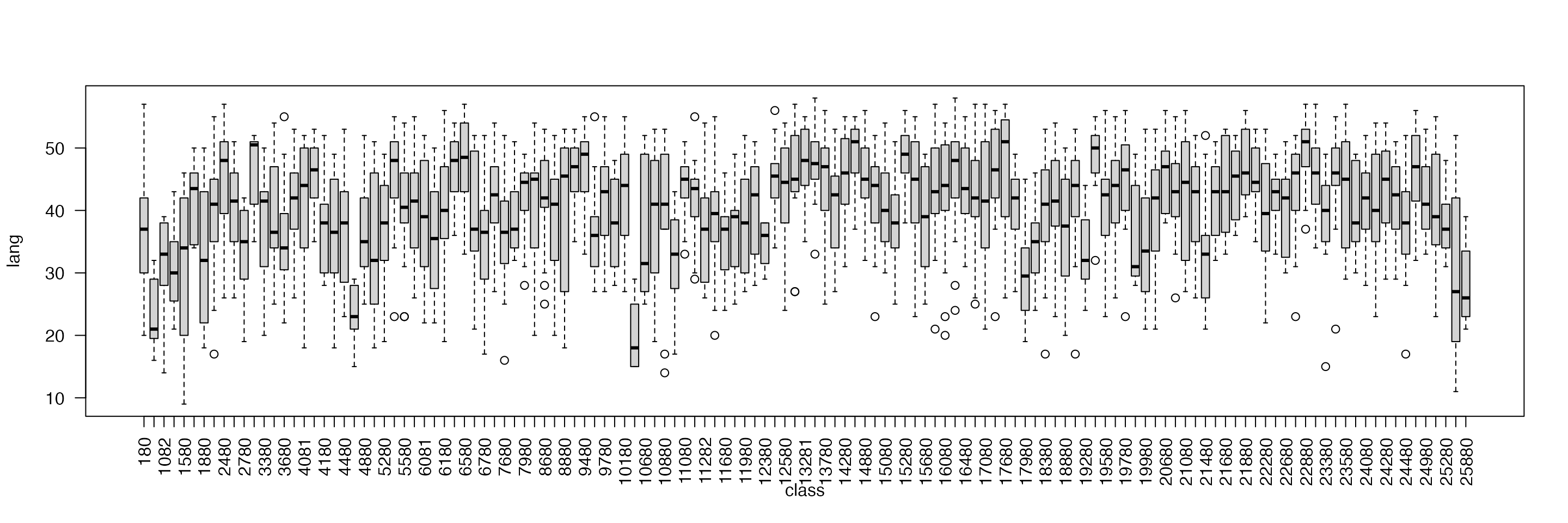
The code to fit the model with random effects is:
## Call:
## bayesx(formula = lang ~ IQ + GS + SES + COMB + sx(class, bs = "re"),
## data = nlschools)
##
## Fixed effects estimation results:
##
## Parametric coefficients:
## Mean Sd 2.5% 50% 97.5%
## (Intercept) 10.6158 1.4532 7.8300 10.5345 13.5842
## IQ 2.2460 0.0749 2.1000 2.2481 2.3950
## GS -0.0154 0.0477 -0.1069 -0.0162 0.0746
## SES 0.1652 0.0149 0.1359 0.1653 0.1966
## COMB1 -1.9744 0.5852 -3.1245 -1.9681 -0.8178
##
## Random effects variances:
## Mean Sd 2.5% 50% 97.5% Min Max
## sx(class):re 8.6044 1.3576 6.1081 8.5338 11.4041 5.0351 14.372
##
## Scale estimate:
## Mean Sd 2.5% 50% 97.5%
## Sigma2 40.0422 1.1947 37.7744 40.0373 42.308
##
## N = 2287 burnin = 2000 method = MCMC family = gaussian
## iterations = 12000 step = 10You can compare this Bayesian fit with a non-Bayesian analysis, this
time making use of the lmer() function in the package
lme4 (here, lme means linear mixed
effects). We present the code only for comparison, and we do not
describe lmer() or lme4 more fully.
## Loading required package: Matrix##
## Attaching package: 'lme4'## The following object is masked from 'package:nlme':
##
## lmList
m2lmer <- lmer(
lang ~ IQ + GS + SES + COMB + (1 | class), # Fit a separate intercept for each level of class
data = nlschools
)
summary(m2lmer)## Linear mixed model fit by REML ['lmerMod']
## Formula: lang ~ IQ + GS + SES + COMB + (1 | class)
## Data: nlschools
##
## REML criterion at convergence: 15132.9
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.9588 -0.6488 0.0486 0.7204 3.0765
##
## Random effects:
## Groups Name Variance Std.Dev.
## class (Intercept) 8.507 2.917
## Residual 39.998 6.324
## Number of obs: 2287, groups: class, 133
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 10.63524 1.50012 7.090
## IQ 2.24696 0.07133 31.502
## GS -0.01562 0.04770 -0.327
## SES 0.16538 0.01479 11.185
## COMB1 -2.02161 0.60055 -3.366
##
## Correlation of Fixed Effects:
## (Intr) IQ GS SES
## IQ -0.484
## GS -0.798 -0.002
## SES -0.062 -0.292 -0.057
## COMB1 -0.168 0.033 -0.006 0.017Generalised Linear Mixed Models
Mixed effects models can also be considered within the context of generalised linear models. In this case, the linear predictor of observation , , can be defined as
Compared to the previous setting of linear mixed effects models, note that now the distribution of the response could be other than Gaussian and that observations are not necessarily nested within groups.
Poisson regression
In this practical we will use the North Carolina Sudden Infant Death
Syndrome (SIDS) data set. It is available in the spData
package and it can be loaded using:
## CNTY.ID BIR74 SID74 NWBIR74
## Min. :1825 Min. : 248 Min. : 0.00 Min. : 1.0
## 1st Qu.:1902 1st Qu.: 1077 1st Qu.: 2.00 1st Qu.: 190.0
## Median :1982 Median : 2180 Median : 4.00 Median : 697.5
## Mean :1986 Mean : 3300 Mean : 6.67 Mean :1051.0
## 3rd Qu.:2067 3rd Qu.: 3936 3rd Qu.: 8.25 3rd Qu.:1168.5
## Max. :2241 Max. :21588 Max. :44.00 Max. :8027.0
## BIR79 SID79 NWBIR79 east
## Min. : 319 Min. : 0.00 Min. : 3.0 Min. : 19.0
## 1st Qu.: 1336 1st Qu.: 2.00 1st Qu.: 250.5 1st Qu.:178.8
## Median : 2636 Median : 5.00 Median : 874.5 Median :285.0
## Mean : 4224 Mean : 8.36 Mean : 1352.8 Mean :271.3
## 3rd Qu.: 4889 3rd Qu.:10.25 3rd Qu.: 1406.8 3rd Qu.:361.2
## Max. :30757 Max. :57.00 Max. :11631.0 Max. :482.0
## north x y lon
## Min. : 6.0 Min. :-328.04 Min. :3757 Min. :-84.08
## 1st Qu.: 97.0 1st Qu.: -60.55 1st Qu.:3920 1st Qu.:-81.20
## Median :125.5 Median : 114.38 Median :3963 Median :-79.26
## Mean :122.1 Mean : 91.46 Mean :3953 Mean :-79.51
## 3rd Qu.:151.5 3rd Qu.: 240.03 3rd Qu.:4000 3rd Qu.:-77.87
## Max. :182.0 Max. : 439.65 Max. :4060 Max. :-75.67
## lat L.id M.id
## Min. :33.92 Min. :1.00 Min. :1.00
## 1st Qu.:35.26 1st Qu.:1.00 1st Qu.:2.00
## Median :35.68 Median :2.00 Median :3.00
## Mean :35.62 Mean :2.12 Mean :2.67
## 3rd Qu.:36.05 3rd Qu.:3.00 3rd Qu.:3.25
## Max. :36.52 Max. :4.00 Max. :4.00A full description of the data set is provided in the associated
manual page (check with ?nc.sids) but in this practical we
will only consider these variables:
BIR74, number of births (1974-78).SID74, number of SID deaths (1974-78).NWBIR74, number of non-white births (1974-78).
These variables are measured at the county level in North Carolina, of which there are 100.
Because SID74 records the number of SID deaths, the
model is Poisson:
Here, represents the number of cases in county and the mean. In addition, mean will be written as , where is the expected number of cases and the relative risk in county .
The relative risk is often modelled, on the log-scale, to be equal to a linear predictor:
The expected number of cases is computed by multiplying the number of births in county to the overall mortality rate
where represents the total number of births in country . Hence, the expected number of cases in county is .
# Overall mortality rate
r74 <- sum(nc.sids$SID74) / sum(nc.sids$BIR74)
# Expected cases
nc.sids$EXP74 <- r74 * nc.sids$BIR74A common measure of relative risk is the standardised mortality ratio ():
nc.sids$SMR74 <- nc.sids$SID74 / nc.sids$EXP74A summary of the SMR can be obtained:
hist(nc.sids$SMR, xlab = "SMR")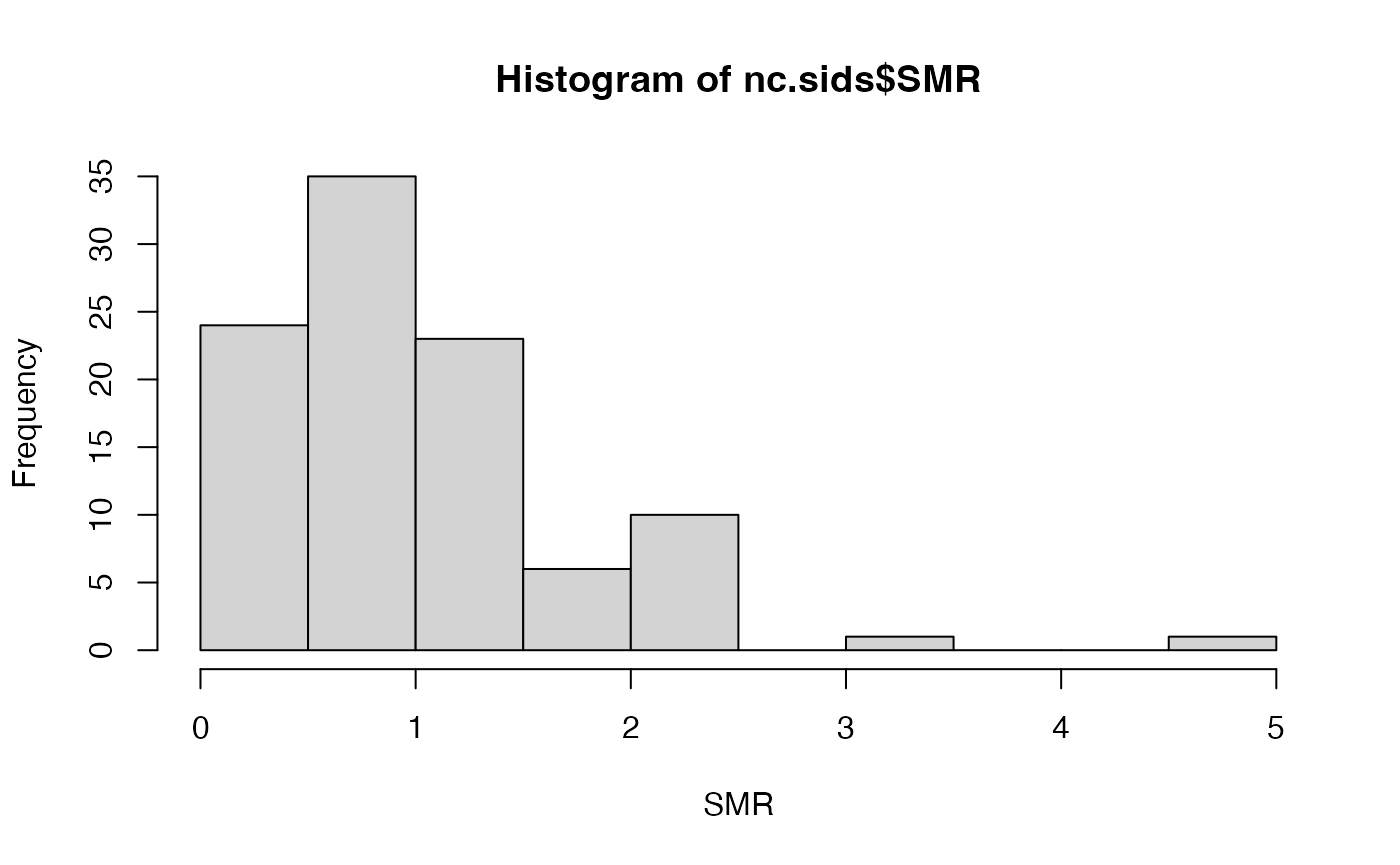
Values above 1 indicate that the county has more observed deaths than expected and that there might be an increased risk in the area.
As a covariate, we will compute the proportion of non-white births:
nc.sids$NWPROP74 <- nc.sids$NWBIR74/ nc.sids$BIR74There is a clear relationship between the SMR and the proportion of non-white births in a county:
plot(nc.sids$NWPROP74, nc.sids$SMR74)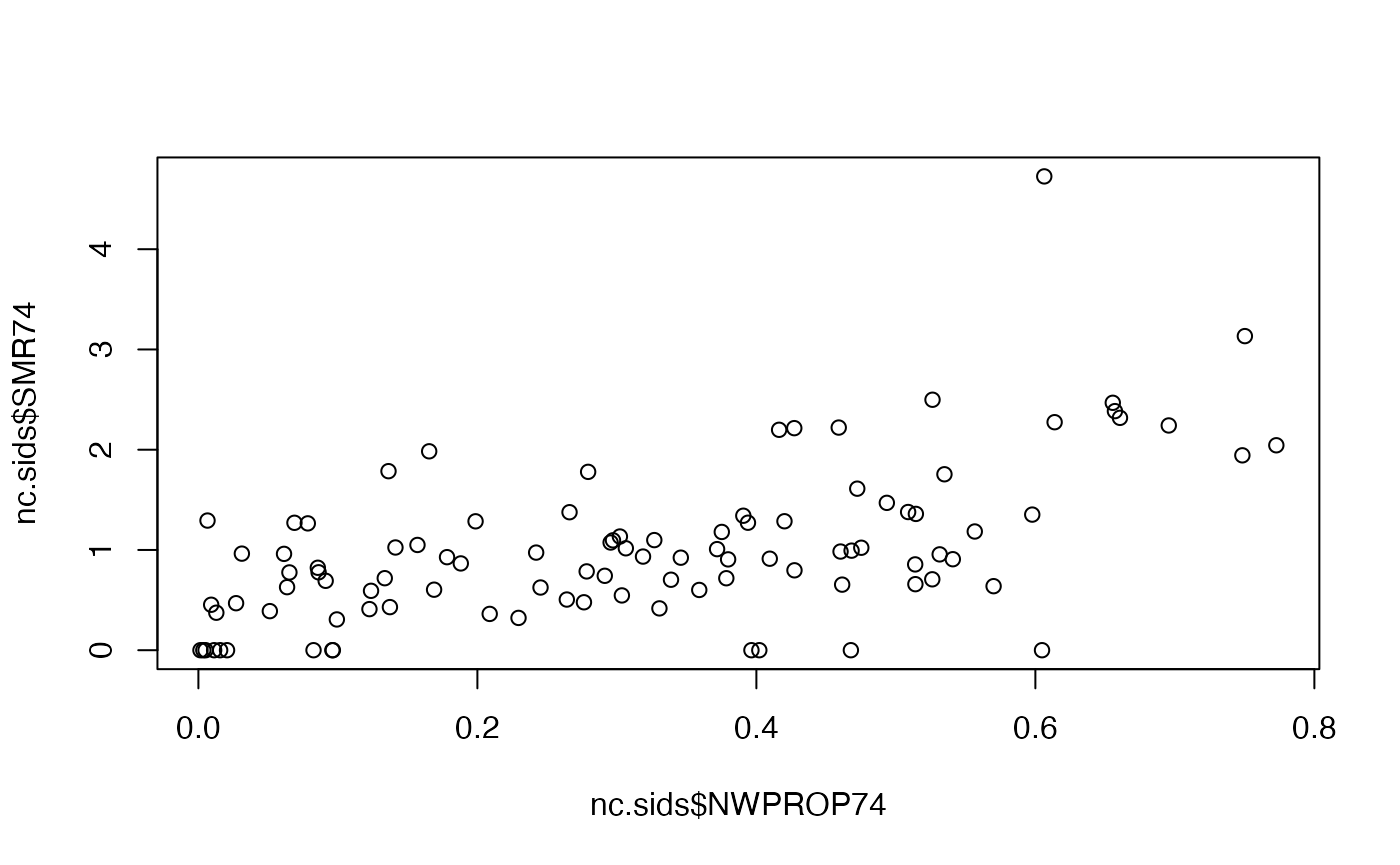
# Correlation
cor(nc.sids$NWPROP74, nc.sids$SMR74)## [1] 0.5793901A simple Poisson regression can be fit by using the following code:
m1nc <- bayesx(
SID74 ~ 1 + NWPROP74,
family = "poisson",
offset = log(nc.sids$EXP74),
data = nc.sids
)
summary(m1nc)## Call:
## bayesx(formula = SID74 ~ 1 + NWPROP74, data = nc.sids, offset = log(nc.sids$EXP74),
## family = "poisson")
##
## Fixed effects estimation results:
##
## Parametric coefficients:
## Mean Sd 2.5% 50% 97.5%
## (Intercept) -0.6463 0.0906 -0.8189 -0.6458 -0.4542
## NWPROP74 1.8710 0.2206 1.4250 1.8681 2.3064
##
## N = 100 burnin = 2000 method = MCMC family = poisson
## iterations = 12000 step = 10Random effects can also be included to account for intrinsic differences between the counties:
# Index for random effects
nc.sids$ID <- seq_len(nrow(nc.sids))
# Model WITH covariate
m2nc <- bayesx(
SID74 ~ 1 + NWPROP74 + sx(ID, bs = "re"),
family = "poisson",
offset = log(nc.sids$EXP74),
data = as.data.frame(nc.sids)
)
summary(m2nc)## Call:
## bayesx(formula = SID74 ~ 1 + NWPROP74 + sx(ID, bs = "re"), data = as.data.frame(nc.sids),
## offset = log(nc.sids$EXP74), family = "poisson")
##
## Fixed effects estimation results:
##
## Parametric coefficients:
## Mean Sd 2.5% 50% 97.5%
## (Intercept) -0.6617 0.1067 -0.8720 -0.6661 -0.4590
## NWPROP74 1.9076 0.2624 1.3904 1.9105 2.4407
##
## Random effects variances:
## Mean Sd 2.5% 50% 97.5% Min Max
## sx(ID):re 0.0571 0.0298 0.0059 0.0551 0.1210 0.0012 0.1709
##
## N = 100 burnin = 2000 method = MCMC family = poisson
## iterations = 12000 step = 10We can see the fitted relationship with NWPROP74:
x.predict <- seq(0,1,length=1000)
y.predict <- exp(coef(m2nc)["(Intercept)","Mean"]+coef(m2nc)["NWPROP74","Mean"]*x.predict)
oldpar <- par(mfrow = c(1, 1))
plot(nc.sids$NWPROP74, nc.sids$SMR74)
lines(x.predict,y.predict)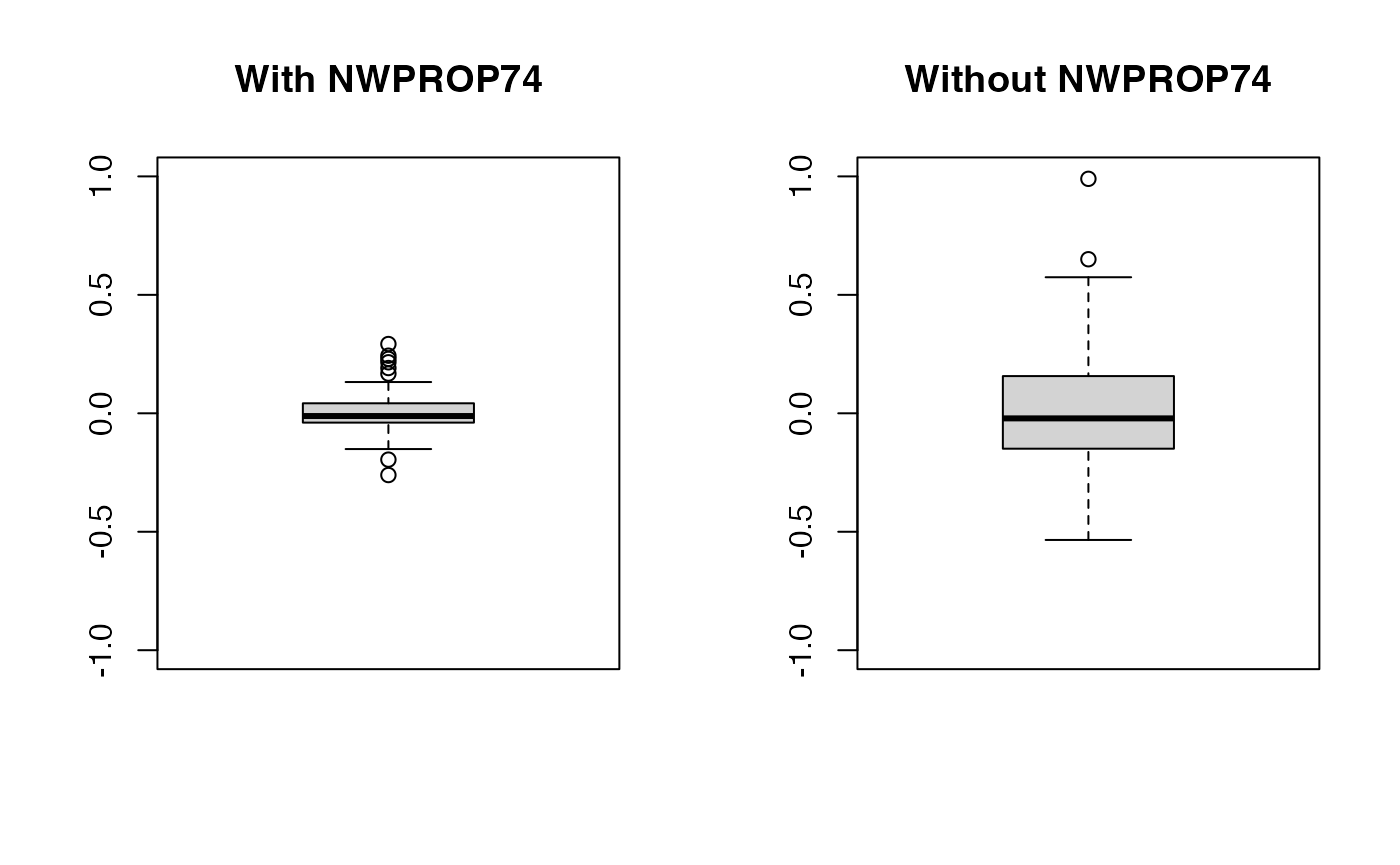
par(oldpar)The role of the covariate can be explored by fitting a model without it:
# Model WITHOUT covariate
m3nc <- bayesx(
SID74 ~ 1 + sx(ID, bs = "re"),
family = "poisson",
offset = log(nc.sids$EXP74),
data = as.data.frame(nc.sids)
)
summary(m3nc)## Call:
## bayesx(formula = SID74 ~ 1 + sx(ID, bs = "re"), data = as.data.frame(nc.sids),
## offset = log(nc.sids$EXP74), family = "poisson")
##
## Fixed effects estimation results:
##
## Parametric coefficients:
## Mean Sd 2.5% 50% 97.5%
## (Intercept) -0.0317 0.0651 -0.1665 -0.0298 0.0864
##
## Random effects variances:
## Mean Sd 2.5% 50% 97.5% Min Max
## sx(ID):re 0.1743 0.0530 0.0914 0.1680 0.2973 0.0454 0.3867
##
## N = 100 burnin = 2000 method = MCMC family = poisson
## iterations = 12000 step = 10Now, notice the decrease in the estimate of the precision of the random effects (i.e., the variance increases). This means that values of the random effects are now larger than in the previous case as the random effects pick some of the effect explained by the covariate.
oldpar <- par(mfrow = c(1, 2))
boxplot(m2nc$effects$`sx(ID):re`$Mean, ylim = c(-1, 1), main = "With NWPROP74")
boxplot(m3nc$effects$`sx(ID):re`$Mean, ylim = c(-1, 1), main = "Without NWPROP74")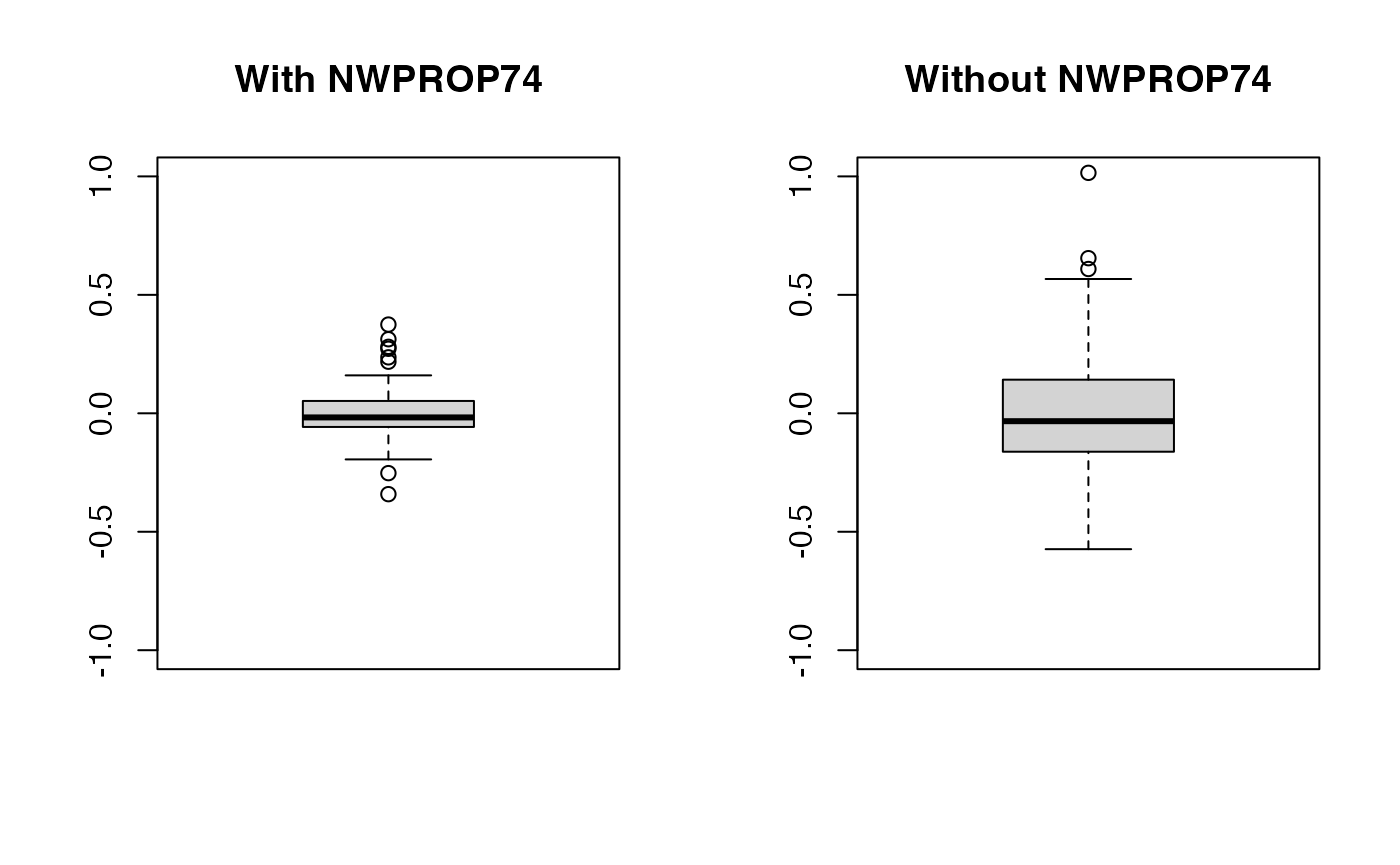
par(oldpar)Further Extensions
Spatial random effects can be defined not to be independent and identically distributed. Instead, spatial or temporal correlation can be considered when defining them. For example, in the North Carolina SIDS data set, it is common to consider that two counties that are neighbours (i.e., share a boundary) will have similar relative risks. This can be taken into account in the model but assuming that the random effects are spatially autocorrelated. This is out of the scope of this introductory course but feel free to ask about this!!
Final Exercises (Optional!!)
We tend to use the final Practical session for catching up and to give you the opportunity to ask us any questions you may have. So please ask away!
If you would like some further exercises to practice, please look at
Practical 8 for more material and examples, including the use of the
non-MCMC package INLA.